Note
Click here to download the full example code
Ramp Analysis¶
Calculate features of Ramp sweeps
Out:
/home/docs/checkouts/readthedocs.org/user_builds/ipfx/envs/latest/lib/python3.6/site-packages/hdmf/spec/namespace.py:485: UserWarning: Ignoring cached namespace 'hdmf-common' version 1.1.0 because version 1.3.0 is already loaded.
% (ns['name'], ns['version'], self.__namespaces.get(ns['name'])['version']))
/home/docs/checkouts/readthedocs.org/user_builds/ipfx/envs/latest/lib/python3.6/site-packages/hdmf/spec/namespace.py:485: UserWarning: Ignoring cached namespace 'core' version 2.2.0 because version 2.2.5 is already loaded.
% (ns['name'], ns['version'], self.__namespaces.get(ns['name'])['version']))
import os
import matplotlib.pyplot as plt
import seaborn as sns
from ipfx.epochs import get_stim_epoch
from ipfx.dataset.create import create_ephys_data_set
from ipfx.utilities import drop_failed_sweeps
from ipfx.feature_extractor import (
SpikeFeatureExtractor, SpikeTrainFeatureExtractor
)
from ipfx.stimulus_protocol_analysis import RampAnalysis
# Download and access the experimental data from DANDI archive per instructions in the documentation
# Example below will use an nwb file provided with the package
nwb_file = os.path.join(
os.path.dirname(os.getcwd()),
"data",
"nwb2_H17.03.008.11.03.05.nwb"
)
# Create Ephys Data Set
data_set = create_ephys_data_set(nwb_file=nwb_file)
# Drop failed sweeps: sweeps with incomplete recording or failing QC criteria
drop_failed_sweeps(data_set)
# get sweep table of Ramp sweeps
ramp_table = data_set.filtered_sweep_table(
stimuli=data_set.ontology.ramp_names
)
ramp_sweeps = data_set.sweep_set(ramp_table.sweep_number)
# Select epoch corresponding to the actual recording from the sweeps
# and align sweeps so that the experiment would start at the same time
ramp_sweeps.select_epoch("recording")
ramp_sweeps.align_to_start_of_epoch("experiment")
# Select epoch corresponding to the actual recording from the sweeps
# and align sweeps so that the experiment would start at the same time
ramp_sweeps.select_epoch("recording")
ramp_sweeps.align_to_start_of_epoch("experiment")
# find the start and end time of the stimulus
# (treating the first sweep as representative)
stim_start_index, stim_end_index = get_stim_epoch(ramp_sweeps.i[0])
stim_start_time = ramp_sweeps.t[0][stim_start_index]
stim_end_time = ramp_sweeps.t[0][stim_end_index]
spx = SpikeFeatureExtractor(start=stim_start_time, end=None)
sptfx = SpikeTrainFeatureExtractor(start=stim_start_time, end=None)
# Run the analysis
ramp_analysis = RampAnalysis(spx, sptfx)
results = ramp_analysis.analyze(ramp_sweeps)
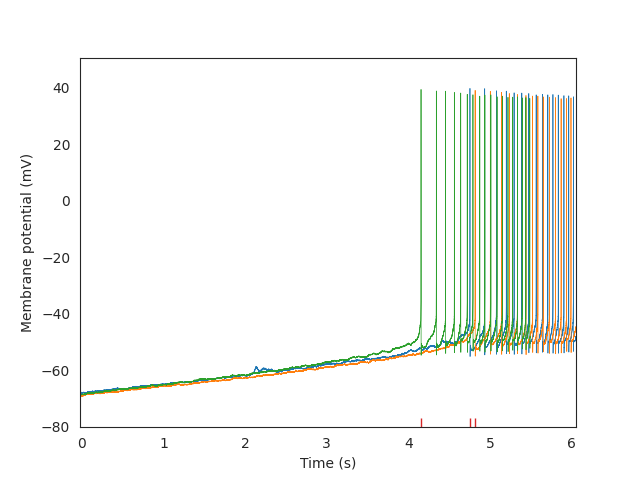
# Plot the sweeps and the latency to the first spike of each
sns.set_style("white")
for swp in ramp_sweeps.sweeps:
plt.plot(swp.t, swp.v, linewidth=0.5)
sns.rugplot(results["spiking_sweeps"]["latency"].values + stim_start_time)
# Set the plot limits to highlight where spikes are and axis labels
plt.xlim(stim_start_time, stim_end_time)
plt.xlabel("Time (s)")
plt.ylabel("Membrane potential (mV)")
plt.show()
Total running time of the script: ( 0 minutes 9.156 seconds)